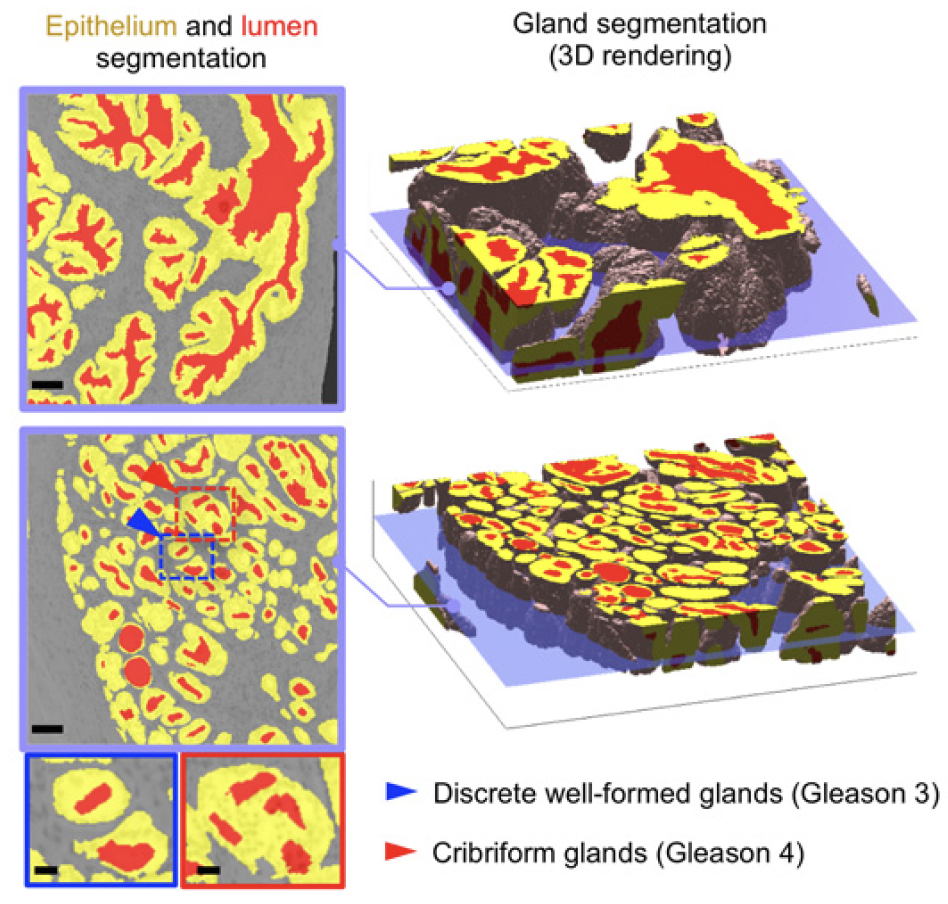
Dataset


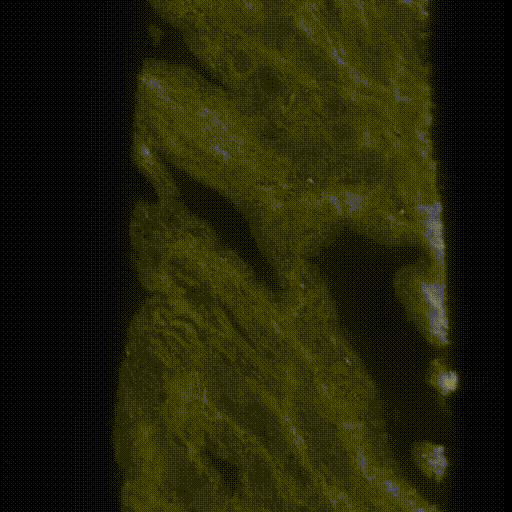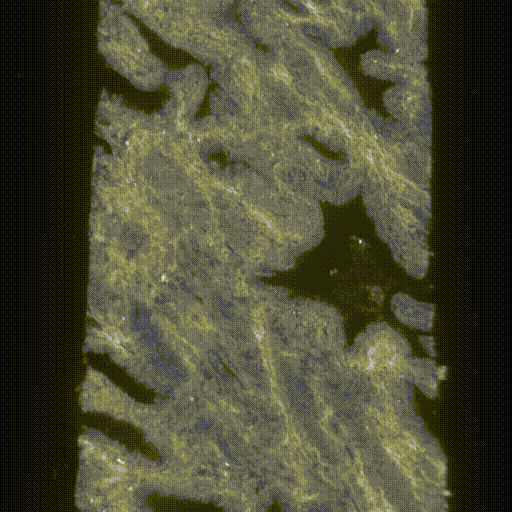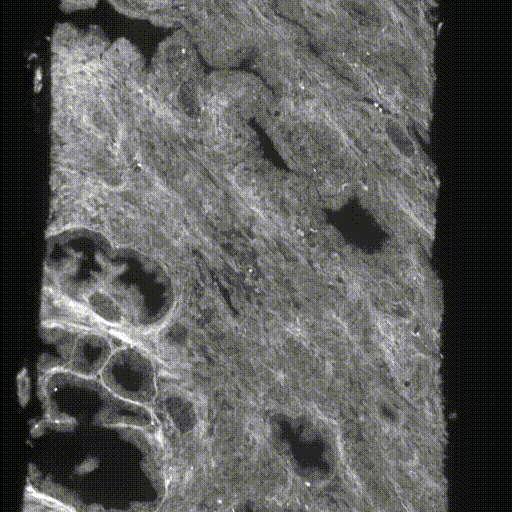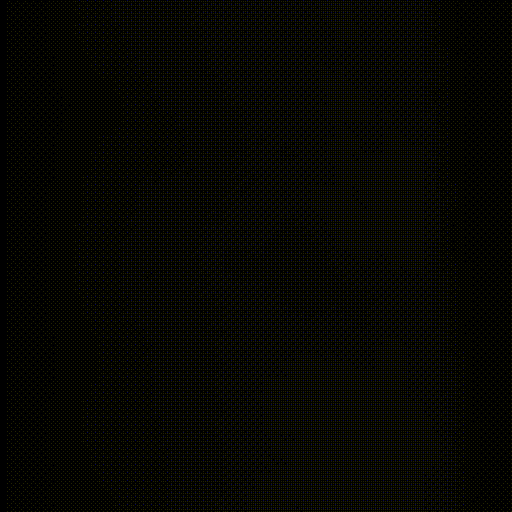
The original data were taken by the lightsheet microscopes in Molecular Biophotonics Laboratory which contains nuclei and cytoplasm channels (or hematoxylin and eosin H&E channels)(Fig.4&5). For memory limit reasons, we use 4x downsampled 3D images (still got ~7000*512*350 in pixel size) for training and inference. There are more than 100 biopsies with ground truth segmentation masks generated previously by traditional thresholding-based CV segmentation methods and verified by senior pathologists (Fig.6). The segmentation masks contain 3 kinds of labels, epithelium, lumen and stroma. We can use those for training in a supervised manner. The inputs are two channels (nuclei and cytoplasm) images and the targets are those corresponding ground truth segmentation masks.