Training
We trained our model with 8 biopsies of data in total (93 blocks, each with 512*512*~350 pixel size). The input data have two channels (nuclei and cytoplasm) and the target (label) data are ground truth segmentation masks with three kinds of labels (epithelium, lumen and stroma). Training on one NVIDIA Quadro P6000 GPU for 700 epochs took ~120 hours (Fig.9). After the model training done, we used it to do inference on another 8 biopsies (115 blocks) of data generated from different cases that are totally unseen by the model, to verify its generalizability.
Results and Discussion
Two pairs of results are randomly selected from the model's inference output. The ones on the left are the ground truth segmentation masks generated by traditional thresholding-based CV methods and were already validated by experienced pathologists. The ones on the right are inference output of the model. The two tiff image stacks (3D images) are from the inference dataset that previously unseen by the model. It turned out that the model can generalize well on unseen images and segment out lumen, epithelium and stroma relatively well. Although some small lumen field were missing or wrongly replaced by epithelium, the result looks good enough given only 8 biopsies of training data with a short training session.




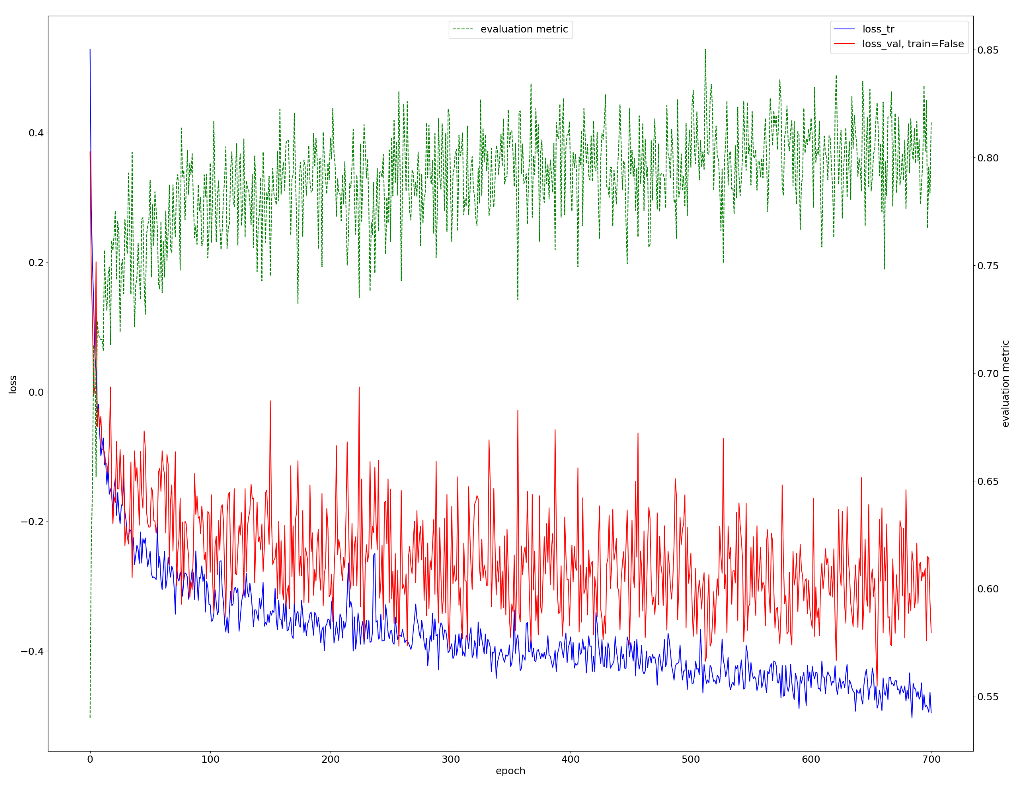
The quantitative evaluation of the training progress with training loss, validation loss and Dice coefficient can be seen from the below image. We can see the loss and evaluation metric fluctuate a lot after ~300 epochs, but it still demonstrates a general trend of decreasing in loss and increasing in performance metric.(Fig.10)
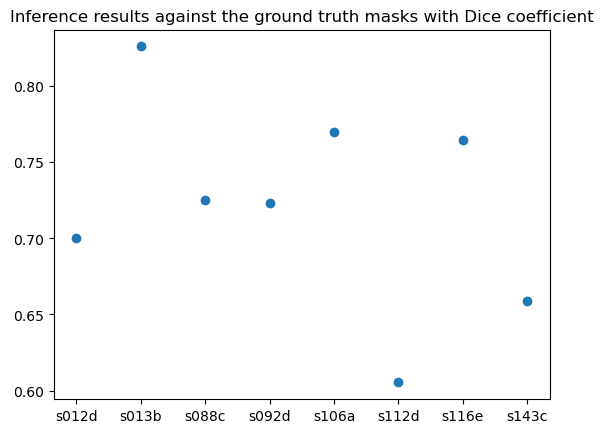
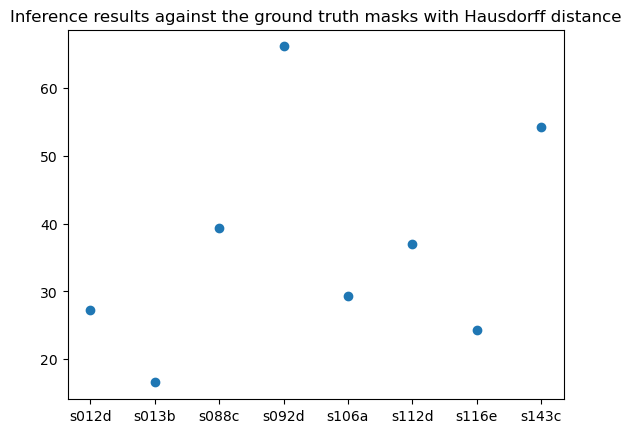
To quantitatively evaluate the model's inference output, we compared the inference results against the ground truth masks with Dice coefficient and Hausdorff distance using the python package “seg-metrics”. The metrics are shown below across all 8 biopsies used for inference. The lowest one got above 0.6 of dice coefficient and the highest got above 0.8, which indicates good performance of the model on test sets.