Abstract
Semantic segmentation is useful to segment out different parts in tissues
from microscopic biomedical images. However, it is a tough task to do traditional
computer vision-based segmentation in 3D since both the continuity and context information need
to be considered for all three directions, many parameters need to be taken into account and much
manual examination and tweaking make it tedious and time-consuming.
Deep learning-based semantic segmentation remains as a promising technology to deal with such problem
since the models generate well on different datasets with similar properties. Once well trained, no more
manual efforts need to be done. There are already a lot of successful deep learning models for specific
segmentation tasks of 3D biomedical images.
nnU-Net is a self-configuring model for various biomedical
segmentation tasks. It can configure different network structures, pre/post-processing pipelines,
etc based on the dataset properties like imaging modality, image sizes, voxel spacings, class
ratios, etc. In this project, we adapt nnU-Net to train a model which is capable of segmenting
out epithelium and lumen of synthetic immunolabeled prostate glands in 3D images.
Problem Statement

Molecular Biophotonics Laboratory
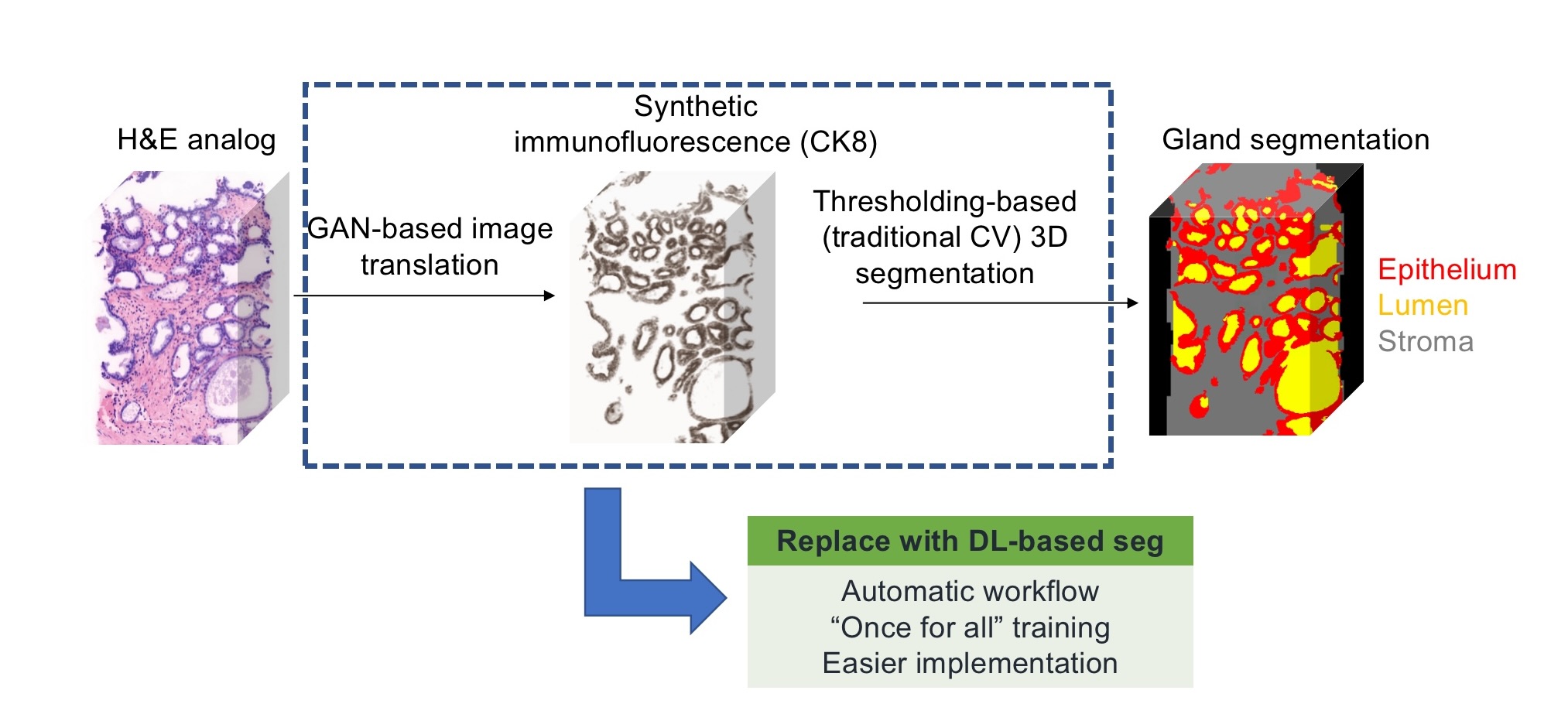
's former member, Dr. Weisi Xie, created an image translation-assisted segmentation in 3D (ITAS3D)
method to segment out different parts (epithelium, lumen, stroma, etc) within 3D images of cancerous prostate
biopsies, thus to help pathologists grade the severity of the cancer (Fig.1). This whole pipeline is great and helpful,
but a bit sophisticated. The final step of the pipeline for gland segmentation is based on traditional computer
vision methods which requires a lot of manual parameter tweaking, we need to do trial-and-errors multiple times
when processing one biopsy, to see the intermediate output and tweak those parameters accordingly for better results,
which is tedious and time-consuming. What's more tedious and time-consuming is that you need to do the manual parameter
tweaking for each and every single biopsy as those parameters don't generate and adapt well for different cases. Thus, we
are always thinking about utilizing deep learning to train another model to automatically do the segmentation.
Our 3D prostate biopsy images are taken by lightsheet microscopes, but major 3D biomedical segmentation models are trained
on CT or MRI images. Transfer learning with those models may require high levels of expertise and experience, with small errors
leading to large drops in performance, some successful configurations from one dataset rarely translate to another.
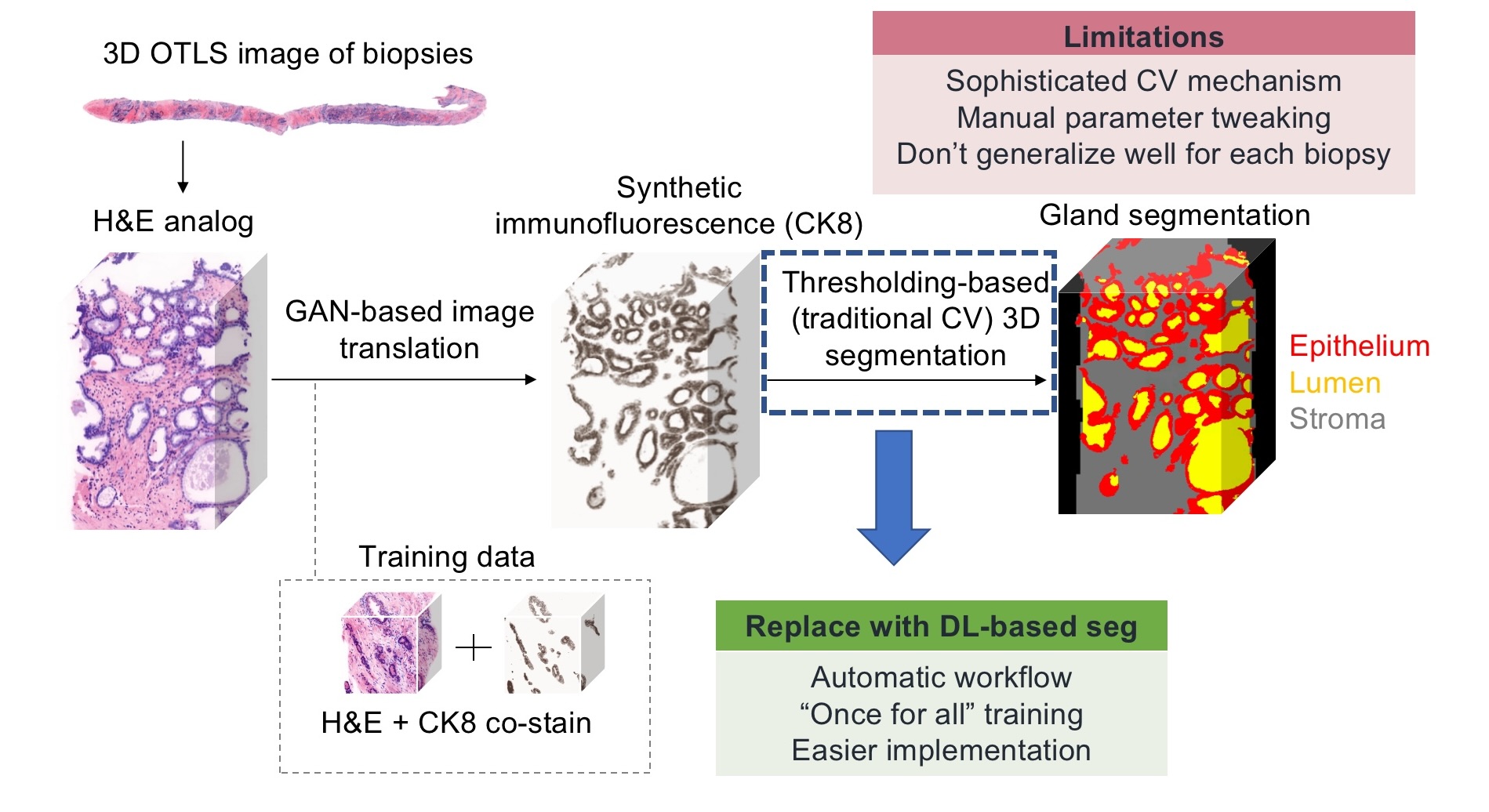
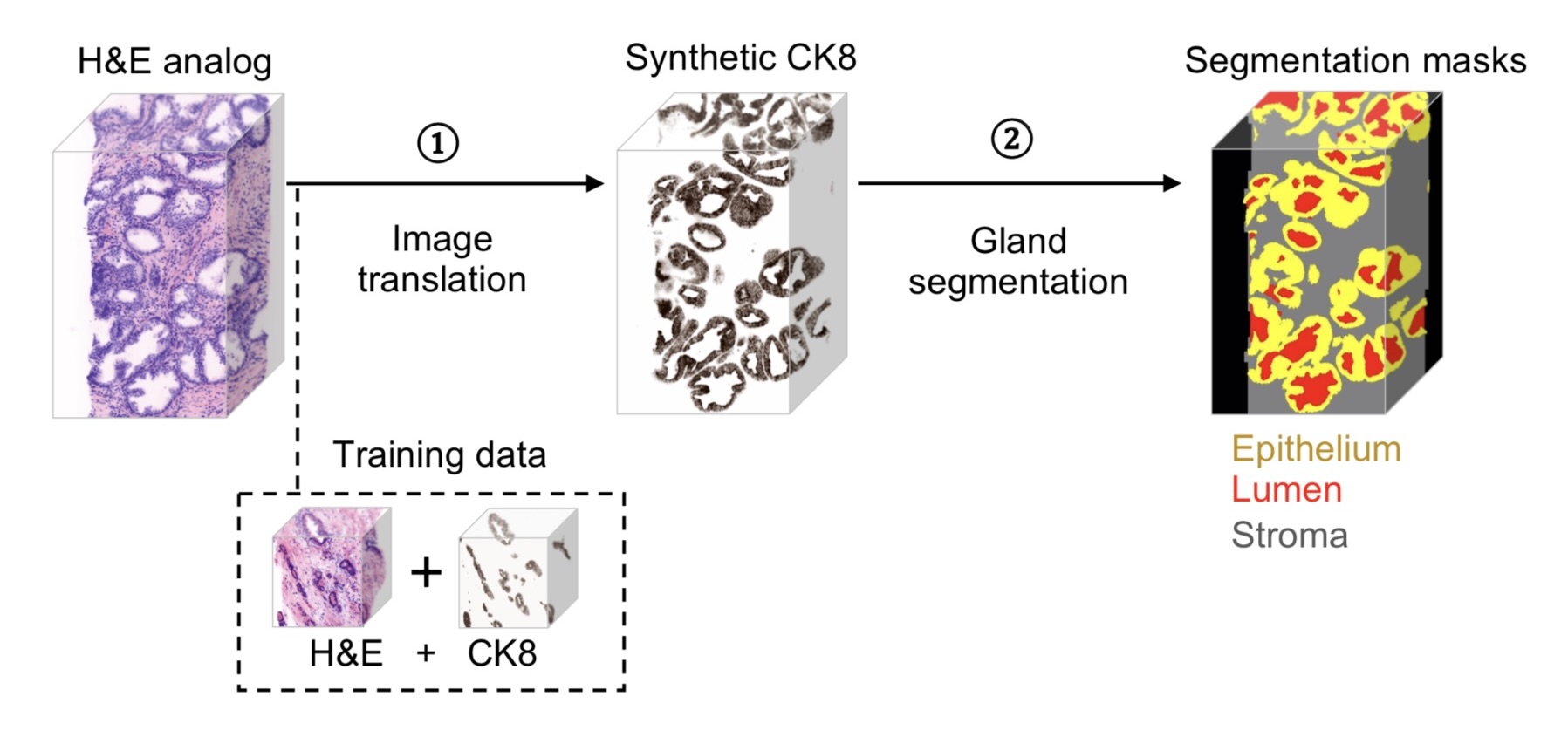
Last quarter, we had successfully done the segmentation based on the synthetic CK8 immunofluorescence 3D images using nnU-Net(Fig.2). Although the synthetic CK8 images, which already have the high contrast and obvious features, are considered easier for segmentation tasks, it does require an additional step of image translation from the H&E channels taken originally from the microscopes. There could be also misleading information generated from this image translation step. In the future work section of our last quarter's project, we said “Eventually, we want to train a model that can directly segment out different parts of tissues from the original nuclei and cytoplasm channels taken from the microscopes.” So this time, we are focused to address this issue, together with some other improvements like adding one stroma mask during segmentation(Fig.3).