Previous Work

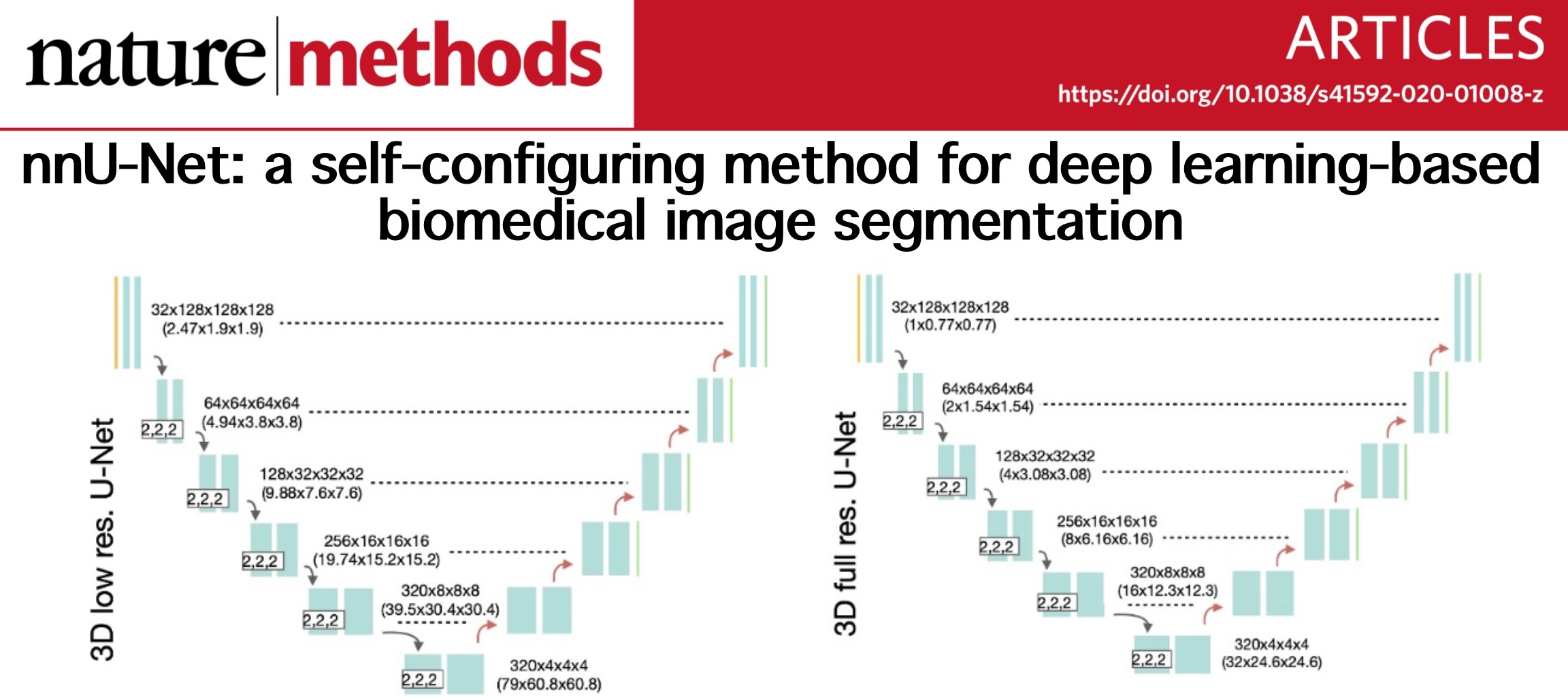
nnU-Net is designed to deal with the dataset diversity found in the biomedical imaging domain. It condenses and automates the key decisions for designing a successful segmentation pipeline for any given dataset. The backbone of nnU-Net, is still composed by several U-Net-like network structures. We know that current biomedical image segmentation models are more or less based on U-Net, a classical network structure for these specific tasks. But nnU-Net can configure suitable network structure, training and pre/post-processing itself for various input dataset. Also, as stated above, the segmentation masks, which we used as ground truth, are previously generated by Dr. Weisi Xie, through ITAS3D pipeline. She did image translation using GAN to generate the synthetic CK8 images from original H&E images taken from our 3D lightsheet microscopes, and then employed traditional thresholding-based segmentation to segment out those epithelium, lumen and stroma parts inside the prostate glands. After considerable parameter tweaking and fine-tuning, all those masks are finally verified by senior pathologists so we can safely use them as ground truth labels to train our nnU-Net model in a supervised learning fashion.
Our Approach
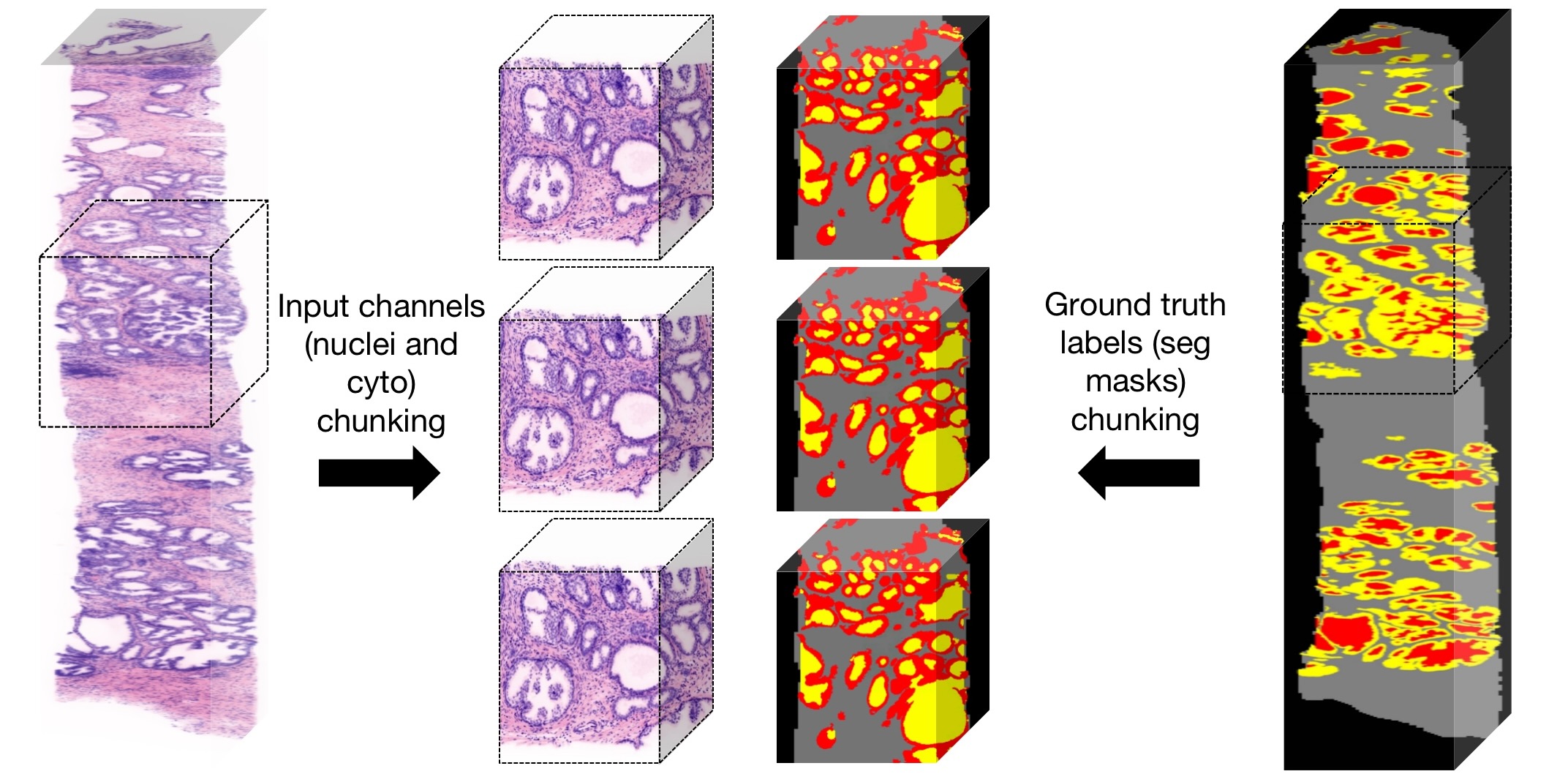
We employed the nnU-Net repository and follow the instruction to install the whole framework for our use. We wrote code to prepare the data and restructure them to the correct format and arrangement for training and inference. Our 3D image data are still very big after 4x downsample (~7000*512*350 in pixel size) thus cannot be directly fed into the network due to memory limit and time concern. Hence, we chunked our data into ~512*512*350 blocks from 4 entire biopsies to form the training and inference dataset(Fig.8). This ensured that no out of memory issue would occur during training and inference, as well as guaranteeing a reasonable training time so we could get some results before the project due, but it will impact on the model's prediction accuracy since some context information would be lost after chunking the image data. The model wouldn't be able to “see” the entire biopsy as a whole while training and might interpret some lumen field as stroma. We also wrote code to separate the nuclei and eosin channels and put them as two different imaging modalities so that the nnU-Net model can take them in for training and inference. The evaluation of the model is integrated in the training process using Dice coefficient as the performance metric. We also examined the segmentation results generated by the trained model after inference, both by visually evaluate how much difference against the ground truth masks qualitatively, and by quantitatively measurements using Dice coefficient and Hausdorff distance 95% percentile.